By the 1980s, when Tom was splashing in a pond by his house catching the frogs, snakes, and salamanders that he’s always loved, biologists had moved beyond Ensatina‘s morphology. New tools allowed researchers2 to study the salamanders’ proteins, the chemical building blocks of life — and what they found supported Robert Stebbins’ ideas. Even though eschscholtzii and klauberi live in the same places in Southern California, their proteins are quite different from one another and each is more biochemically similar to its northern neighbor. This is exactly what we’d expect to observe if these two subspecies represent the endpoints of the ring species. Furthermore, populations of picta had great variety in their proteins — and that’s typically what we observe in populations that have been established for a long time and have had plenty of time to accumulate mutations. This observation supported the idea that picta‘s immediate ancestors gave rise to the whole ring species.
By the 1990s, when Tom was in high school considering a career in biology, technology had progressed even further. Scientists could now study the sequence of DNA inside mitochondria — the cellular organelles that help power cells. Ensatina‘s mitochondrial DNA3 showed the same patterns as their proteins: the northern salamander lineages had the greatest variety of sequences, suggesting that Ensatina got its start in the north and moved south through California.
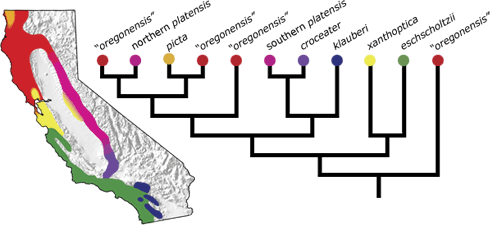
Biologists also used mitochondrial DNA sequences to try to figure out the family tree for all the salamander subspecies. They found that the northern salamander lineages branched off near the base of that family tree — suggesting that they are closely related to the ancestor of the ring. The forms near the eastern and western endpoints of the ring formed distinct clans just as we’d expect if they each evolved separately from Ensatina‘s ancestor, as proposed by Robert.
Get tips for using research profiles, like this one, with your students.
2Wake, D.B., and K.P. Yanev. 1986. Geographic variation in allozymes in a "ring species," the plethodontid salamander Ensatina eschscholtzii of western North America. Evolution 40:702-715.
3Moritz, C., C.J. Schneider, and D.B. Wake. 1992. Evolutionary relationships within the Ensatina eschscholtzii complex confirm the ring species interpretation. Systematic Zoology 41:273-291.